Analysis of ubiquition by synthetic biology
The study of ubiquitinated proteins faces many challenges such as the low stoichiometry of ubiquitin, rapid cleavage of ubiquitin moieties and the degradation of modified proteins. By reconstituting the ubiquitination cascade in an orthogonal systems such as E. coli, we are able to circumvents some of these limitations. For this purpose we developed a synthetic biology toolbox that is based on golden gate cloning (Kowarschik et al.). Expression operons containing all components of the ubiquitination cascade, enable the easy analysis E3 activity or substrate ubiquitination, which can be expensive and time-consuming. Furthermore, UbiGate is also amenable for easy up-scaling, rendering ubiquitinated products accessible to further down-stream analysis such as mass-spectrometry or NMR.
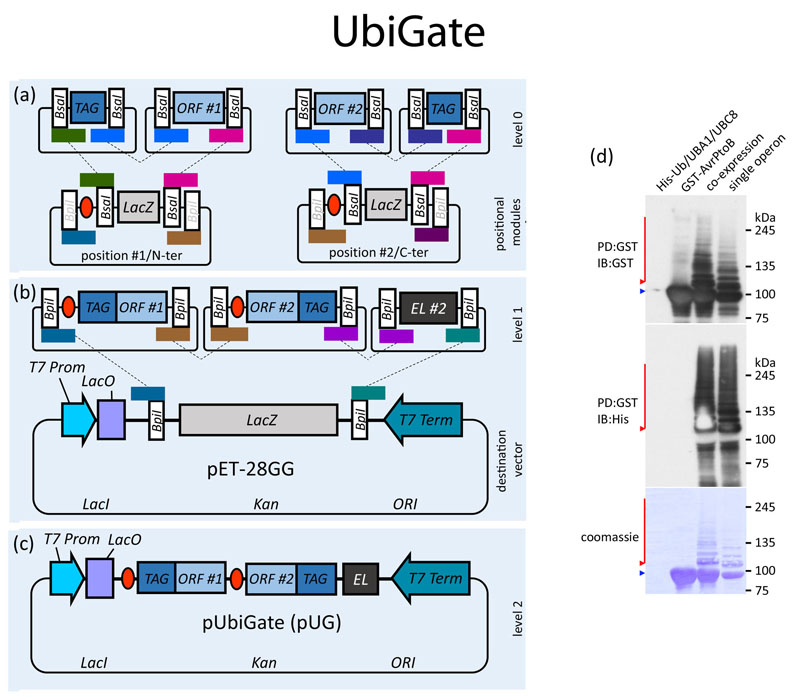
UbiGate assembly of polycistronic operons for ubiquitination analysis.
A) Open reading frames (ORFs) amplified by PCR with flanking BpiI recognition sites are cloned by restriction-ligation into level 0 vectors (only the resulting level 0 constructs containing an insert are shown). Level 0 vectors containing an ORF, and optionally a tag, are cloned into level 1 positional vectors using BsaI. The four nucleotide unique cleavage overhangs (UCOs) resulting from type IIS restriction are depicted as coloured boxes over BpiI or BsaI recognition sites. Note that UCOs are generated in distinct orientations in dependence of recognition site orientation. Two sets of six level 1 positional vectors allow cloning of ORFs with N- and C-terminal tags for six defined positions. B) The destination vector pET-28 Golden-Gate (pET-28GG) was created based on the pET-28b vector containing a LacZ flanked by BpiI sites for blue/white selection. C) Level 1 vectors are assembled into the pET-28GG by BpiI restriction-ligation, resulting in the desired level 2 polycistronic operon. D) Large amounts of ubiquitinated proteins can be purified immobilized metal ion chromatography (IMAC) using the His-ubiquitin.
All materials are available at Addgene
